Official Pytorch Code base for UNeXt: MLP-based Rapid Medical Image Segmentation Network, MICCAI 2022
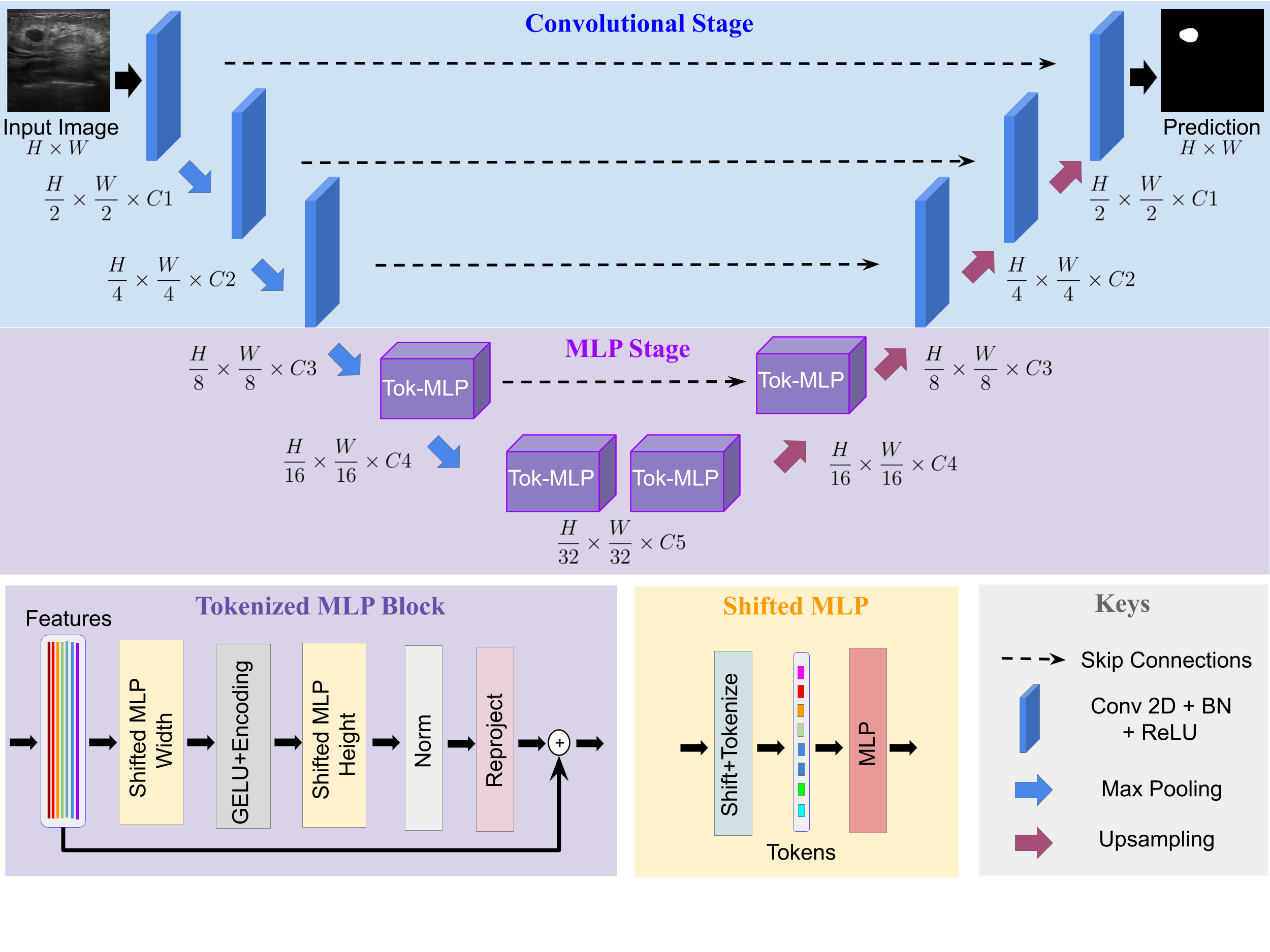
UNet and its latest extensions like TransUNet have been the leading medical image segmentation methods in recent years. However, these networks cannot be effectively adopted for rapid image segmentation in point-of-care applications as they are parameter-heavy, computationally complex and slow to use. To this end, we propose UNeXt which is a Convolutional multilayer perceptron (MLP) based network for image segmentation. We design UNeXt in an effective way with an early convolutional stage and a MLP stage in the latent stage. We propose a tokenized MLP block where we efficiently tokenize and project the convolutional features and use MLPs to model the representation. To further boost the performance, we propose shifting the channels of the inputs while feeding in to MLPs so as to focus on learning local dependencies. Using tokenized MLPs in latent space reduces the number of parameters and computational complexity while being able to result in a better representation to help segmentation. The network also consists of skip connections between various levels of encoder and decoder. We test UNeXt on multiple medical image segmentation datasets and show that we reduce the number of parameters by 72x, decrease the computational complexity by 68x, and improve the inference speed by 10x while also obtaining better segmentation performance over the state-of-the-art medical image segmentation architectures.
The code is stable while using Python 3.6.13, CUDA >=10.1
- Clone this repository:
git clone https://github.com/jeya-maria-jose/UNeXt-pytorch
cd UNeXt-pytorchTo install all the dependencies using conda:
conda env create -f environment.yml
conda activate unextIf you prefer pip, install following versions:
timm==0.3.2
mmcv-full==1.2.7
torch==1.7.1
torchvision==0.8.2
opencv-python==4.5.1.48Make sure to put the files as the following structure (e.g. the number of classes is 2):
inputs
└── <dataset name>
├── images
| ├── 001.png
│ ├── 002.png
│ ├── 003.png
│ ├── ...
|
└── masks
├── 0
| ├── 001.png
| ├── 002.png
| ├── 003.png
| ├── ...
|
└── 1
├── 001.png
├── 002.png
├── 003.png
├── ...
For binary segmentation problems, just use folder 0.
- Train the model.
python train.py --dataset <dataset name> --arch UNext --name <exp name> --img_ext .png --mask_ext .png --lr 0.0001 --epochs 500 --input_w 512 --input_h 512 --b 8
- Evaluate.
python val.py --name <exp name>
This code-base uses certain code-blocks and helper functions from UNet++, Segformer, and AS-MLP. Naming credits to Poojan.
@article{valanarasu2022unext,
title={UNeXt: MLP-based Rapid Medical Image Segmentation Network},
author={Valanarasu, Jeya Maria Jose and Patel, Vishal M},
journal={arXiv preprint arXiv:2203.04967},
year={2022}
}